EpiCypher新品促销—— CUTANA™ CUT&Tag Kit升级版(V 2)

EpiCypher CUTANA™ CUT&Tag Kit升级版(Version 2)来了!升级版各方面表现都优于V1版本,
助您持续生成高质量的CUT&Tag数据。新品推广期间凡订购EpiCypher CUTANA™ CUT&Tag Kit,可享额外20%折扣优惠!
前30名下单的客户,还有好礼相送(加油鸭挂件一个)。
促销时间:即日起-2024.9.25
促销代码:CTK20
【 促销产品清单 】
|
供应商 |
产品货号 |
产品名称 |
产品规格 |
|
EpiCypher |
14-1102 |
CUTANA™ CUT&Tag Kit with Primer Set 1 |
48 Reactions |
|
EpiCypher |
14-1103 |
CUTANA™ CUT&Tag Kit with Primer Set 2 |
48 Reactions |
【 六个常见问题带您了解CUT&Tag新变革 】

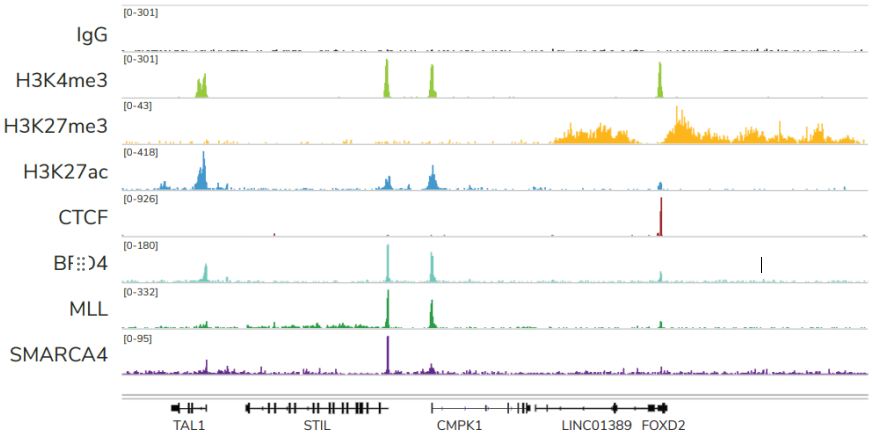
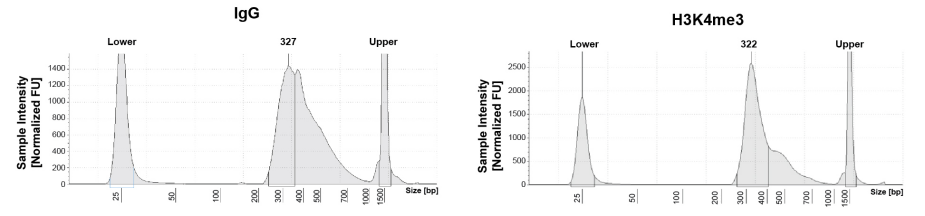
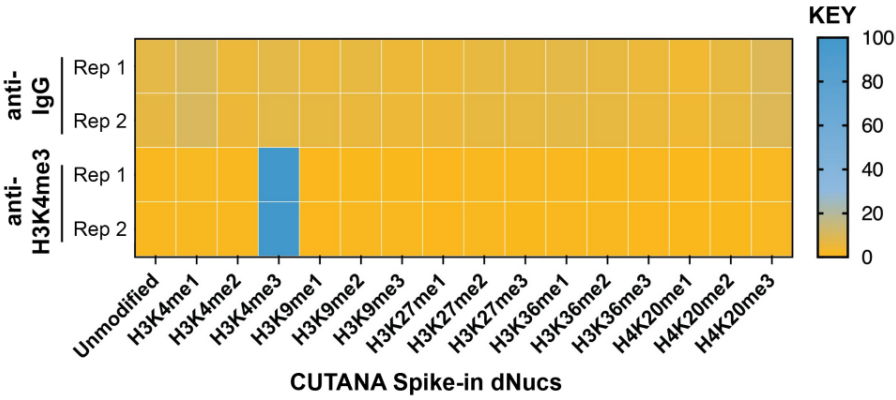
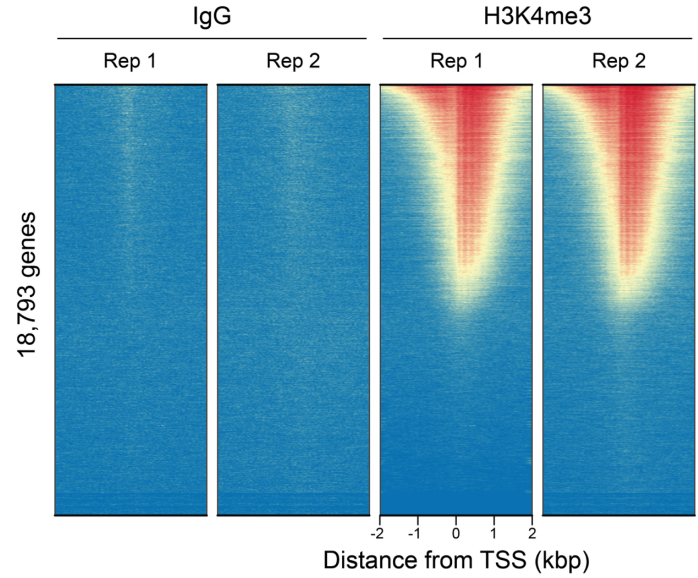
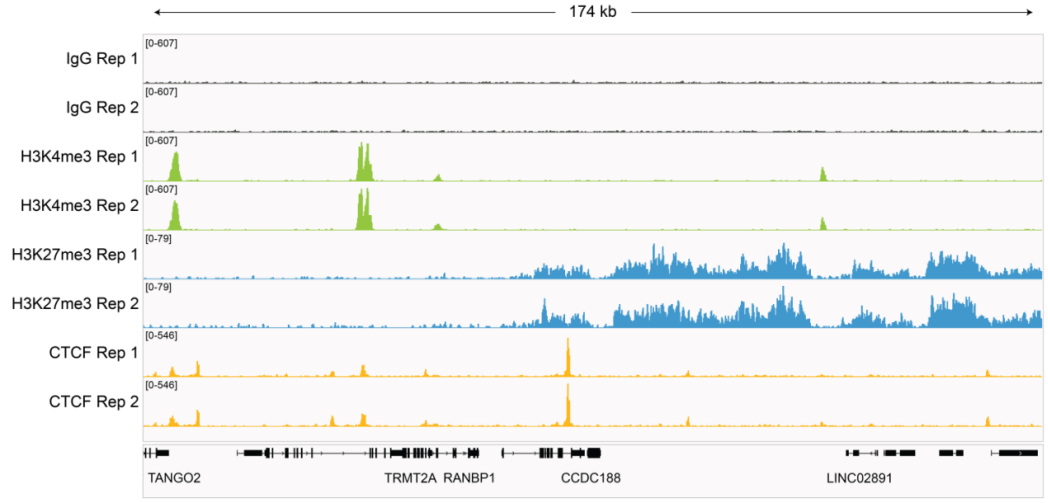
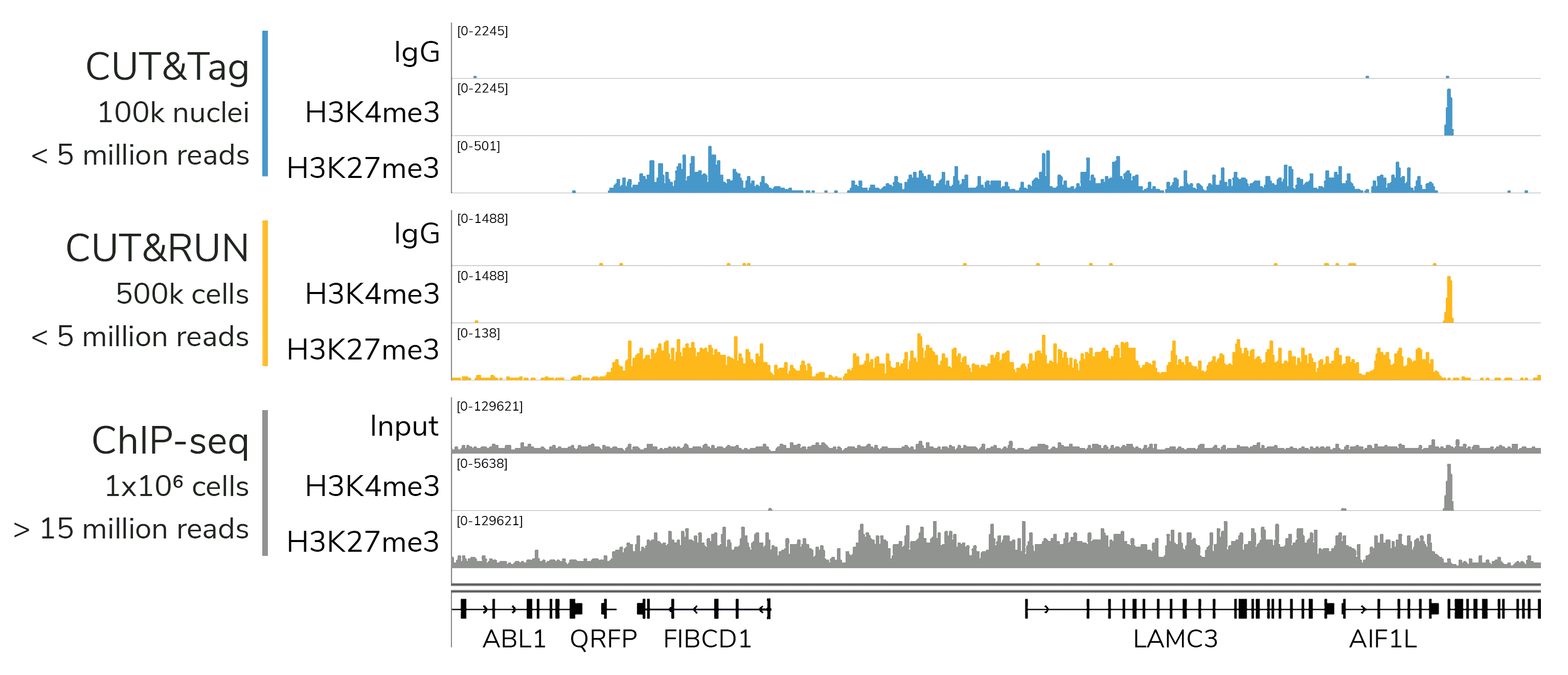
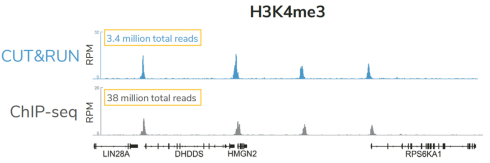
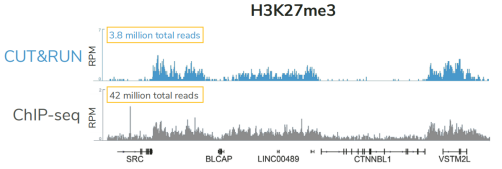
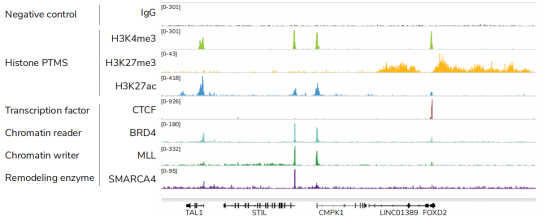
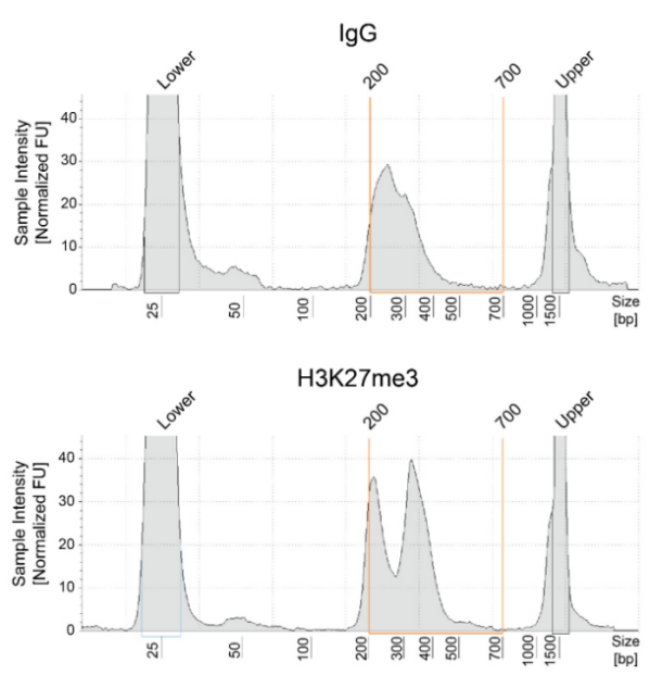
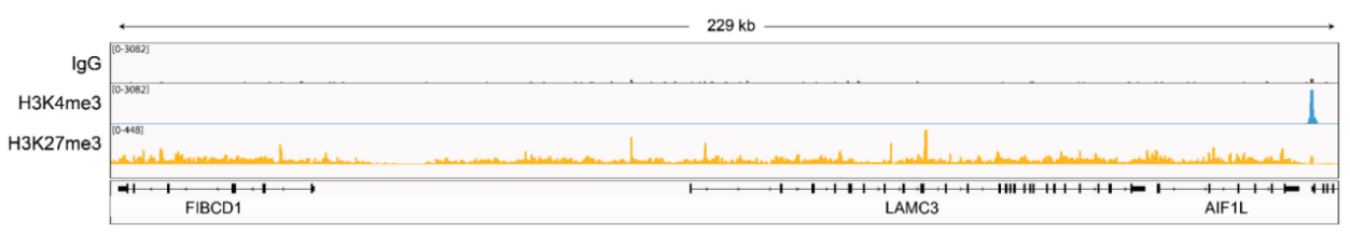
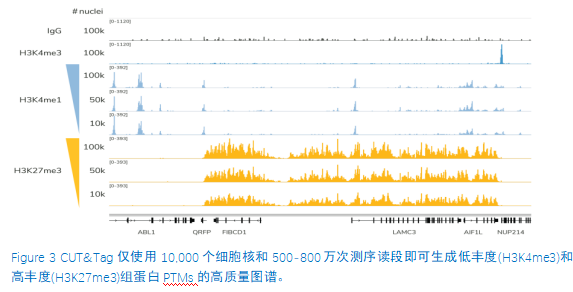
从单细胞图谱分析、空间表观基因组学到 FFPE 样本分析,CUT&Tag正在成为研究热点1-7。 CUT&Tag也是EpiCypher的研发重点。多年来,EpiCypher的团队开发并优化了多种表观基因组图谱检测方法,包括CUT&Tag、CUT&RUN和ChIP-seq。EpiCypher团队充分利用专业知识研发的CUTANA™ CUT&Tag试剂盒,使这项令人难以置信的技术能够走进全球更多的研究领域和实验室。CUTANA™ CUT&Tag 试剂盒第2版现已上市,而且各方面都优于之前的版本。
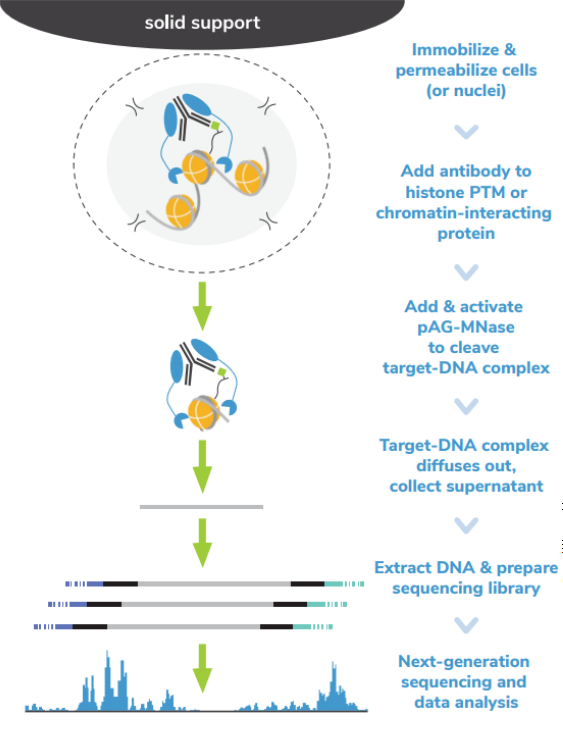
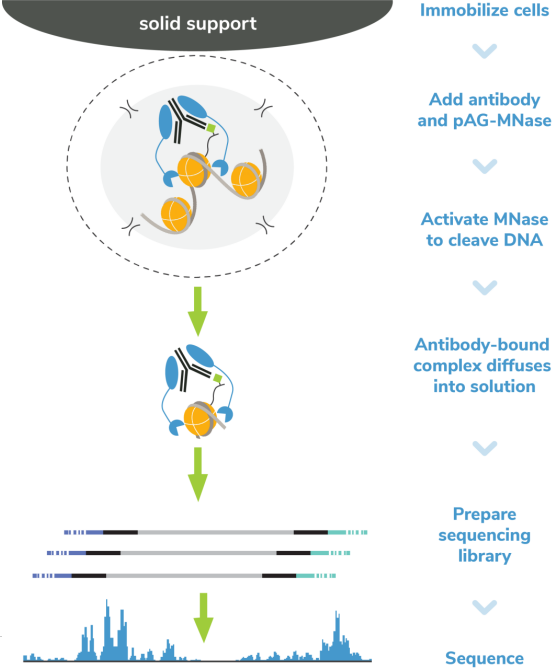
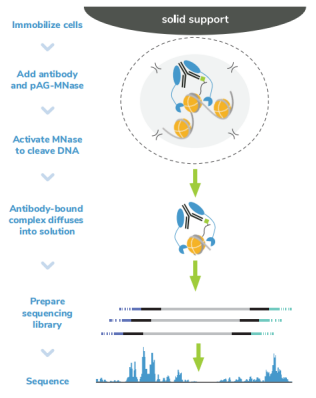
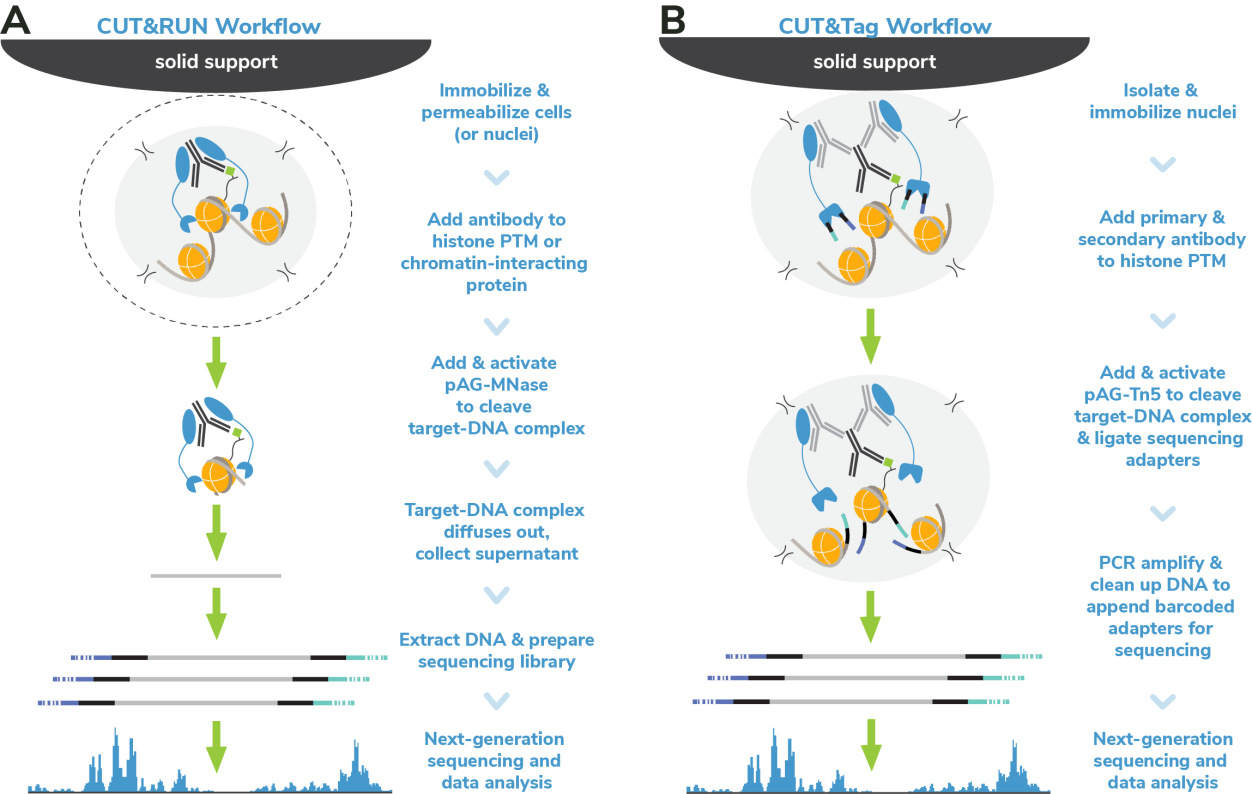
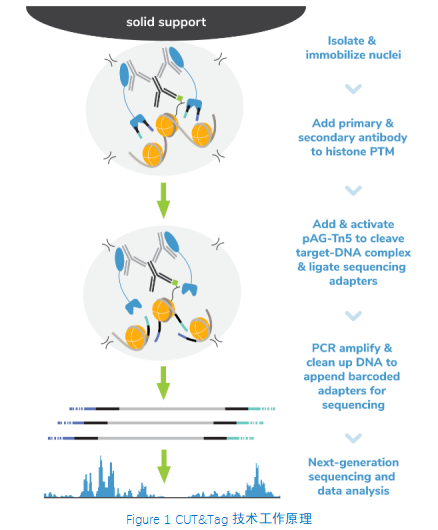
从表面上看,CUT&Tag非常简单8,9。Protein A/G的融合体在抗体标记的染色质上栓了一种过度活跃的Tn5转座酶。激活的Tn5插入接头序列并切割DNA,这一过程被称为标签化。在EpiCypher的Direct-to-PCR方法中,使用识别接头序列的引物从反应混合物中直接扩增标记DNA,绕过文库制备,提高对低细胞数的敏感性。整个测定过程使用的是与磁珠结合的完整细胞核,并对在使用8联排管时的方法进行了优化,从而实现高通量工作流程和自动化解决方案。
为了简化实验过程,EpiCypher团队开发了CUTANA™ CUT&Tag试剂盒,其中包括从细胞到纯化的测序文库所需的所有试剂。请注意,CUT&Tag仅推荐用于组蛋白翻译后修饰(PTMs)的定位。 欲进一步了解如何使用CUT&Tag与其姊妹技术CUT&RUN,请您参阅这篇文章:http://www.jinpanbio.com/xwzx_2376.html。
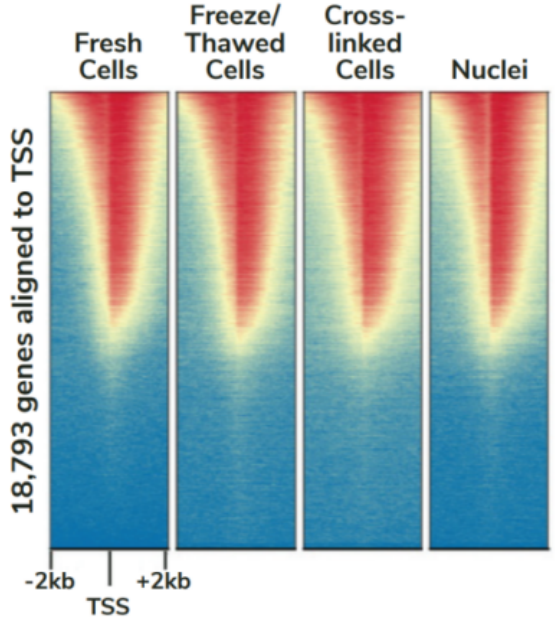
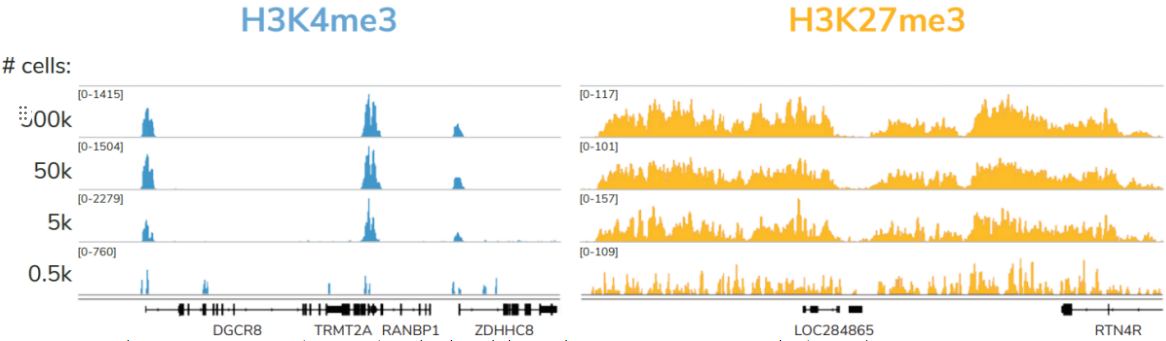
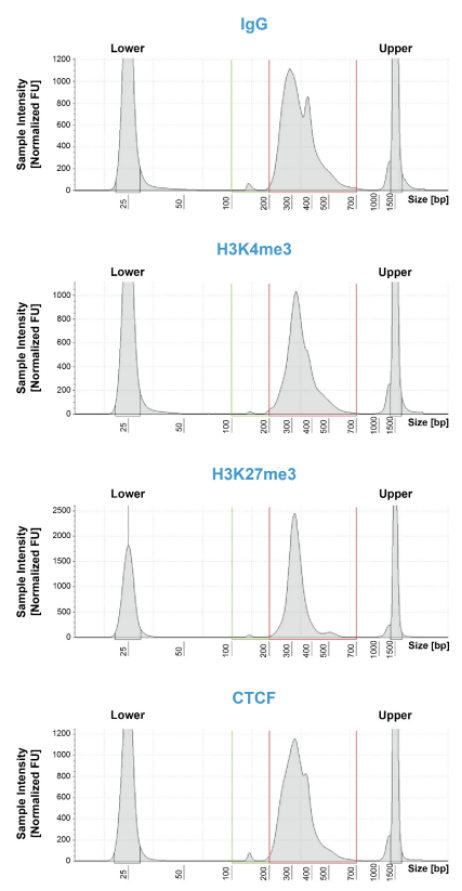
尽管已取得了一些进展,但CUT&Tag仍然是一项具有挑战性的表观基因组检测技术。为了使其方法更加可靠,EpiCypher研发团队不断测试CUTANA™ CUT&Tag实验方案,研究不同的缓冲液成分,调整反应条件,并测试了多种细胞类型。CUTANA™ CUT&Tag试剂盒的第二版展现了EpiCypher最新的优化成果。
✍ 为什么科研学者会对CUT&Tag感兴趣?
科研人员之所以对CUT&Tag非常感兴趣,是因为它可以让科学家在不降低数据质量的情况下简化实验流程,这是ChIP不可能做到的。自从CUT&Tag出现以来,研发人员终于能够开发出更高通量的染色质图谱分析。与ChIP-seq相比,CUT&Tag可以使用更少的细胞,花费更少的时间,对更多的反应进行多重测序,并生成更高分辨率的数据。
✍ 为什么CUT&Tag比ChIP-seq和CUT&RUN快得多呢?
由于各种原因,CUT&Tag比其他染色质图谱分析技术更快。从样本处理的角度看,CUT&Tag无需再多花时间优化细胞裂解、交联或染色质碎裂。CUT&Tag使用完整的细胞核,可在15分钟内从新鲜或冷冻的细胞中获取。然后将细胞核与固体支撑物(ConA磁珠)结合,这样就能限制样品的损失,并可使用磁力架快速清洗。这一特点使该方法与8联排管兼容,进一步加快了实验速度。
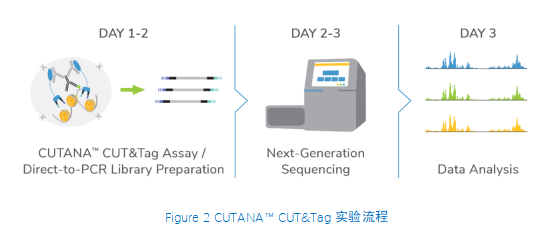
从实验方法的角度看,直接插入测序接头无需进行标准的文库准备步骤,不仅节省了一天的工作时间,而且减少了样品损失。大多数CUT&Tag实验流程中需要纯化DNA,然后进行 PCR,而EpiCypher的Direct-to-PCR策略允许实验人员直接从CUT&Tag反应中扩增标记的DNA。因为所有的实验过程都是在8联排管中进行的,所以实验人员可以将接头引物和PCR混合物直接加入到CUT&Tag反应管中。对于时间紧张的项目,实验人员可以在CUT&Tag第2天结束时加载测序仪,第3天就可以开始处理数据。对于科研人员来讲,这样的周转时间非常宝贵。
✍ 与ChIP-seq相比,为什么在CUT&Tag中可以使用更少的细胞呢?
因为简化了实验流程,降低了背景信号影响,所以CUT&Tag需要的起始细胞数量较少。在 ChIP-seq中,需要交联和染色质超声处理或片段化来制备input的染色质。在免疫沉淀(IP)步骤中,抗体被添加到染色质片段化池中。理想情况下,抗体只与靶标结合,但免疫沉淀几乎总是能回收非靶标片段,并在测序数据中引入背景信号或测序假象。科学家通常会增加细胞数量以克服背景信号影响并提高数据质量,但这种解决方法会限制低数量细胞的应用。
CUT&Tag使用与磁珠结合的完整细胞核,不需要按照传统的IP步骤进行实验,从而减少了背景信号影响。EpiCypher的方法还绕过了ChIP和标准文库制备中的多个DNA纯化步骤,有助于减少样本的损失。总之,与ChIP-seq相比,CUT&Tag可以减少约10倍的材料,这是非常不可思议的。
✍ 科研学者在使用CUT&Tag实验操作流程时最常见的问题是什么呢?
研究者进行CUT&Tag实验时遇到的主要问题是产量低甚至没有产量,这可能是由多种变量影响的。产率低通常是由样品预处理不佳、实验过程中样品损失、ConA磁珠结合不上以及反应混合问题造成的。这些问题都有关联,因此解决起来比较复杂。
样品制备不良的表现是细胞裂解、细胞核制备中有碎片或ConA磁珠结合后上清液中存在未结合的细胞核。如果样本预处理出现上述情况,就可能无法进行标签化,进而导致产量低。
混合不充分也是产量低的一个常见原因。保持磁珠在溶液中对检测的成功至关重要:它有助于确保抗体和pAG-Tn5的均匀分布,并有助于高效的indexing PCR。然而,在实验方案中的第2天,ConA 磁珠浆会变得粘稠且难以重悬,尤其是在标记之后。虽然在处理材料时应当温和,但如果不能很好的混合CUT&Tag反应物,就会严重降低产量。 EpiCypher实验方案详细说明了何时以及如何混合样品,以便最终稳定地回收CUT&Tag生成的文库。
✍ 新版CUT&Tag试剂盒有什么亮点呢?
版本更新的重点是帮助使用者持续生成高质量的CUT&Tag数据。EpiCypher的团队详细讨论了实验流程的每个步骤。比如,为什么缓冲液要使用某种成分或pH值?对细胞核或细胞生理有何影响?这些问题帮助EpiCypher团队找到了可以改进的关键点。
pAG-Tn5的表征表明,酶本身并不是影响产量问题所在,且标记反应是高效的,但样品在标记后的实验步骤中损失了。为此,EpiCypher团队对样品处理、缓冲液成分、方案步骤进行了广泛的头对头比较研究,并对优秀的竞争产品进行了测试。
这些实验揭开了问题的神秘面纱。TAPS Buffer中的低盐浓度导致了渗透性变化和细胞核裂解。混合技术也尤为重要,它是造成样品损失的原因之一。例如,加入SDS Release Buffer后,样品变得粘稠,无法移液。在实验方案的其他部分,由于涡旋使材料粘在离心管边上,也会导致样品损失。
根据实验结果,EpiCypher团队去掉了标记后的低盐 TAPS Buffer洗涤,取而代之的是含有生理盐的Pre-Wash Buffer,以保持细胞核的完整性。EpiCypher团队还完善了实验流程中具体的重悬浮和混合方法,以帮助指导使用者进行最佳操作。EpiCypher内部测试了修改后的CUT&Tag实验方案,结果发现新手和有经验的使用者的实验成功率都有所提高,这说明了CUT&Tag改进后的实用性。
✍ 这些实验操作流程的变化适用于正在使用第1版CUTANA™ CUT&Tag Kit或DIY CUT&Tag的科研人员吗?
是的。所有实验流程的更改都与CUTANATM CUT&Tag Kit的第1版以及 DIY CUT&Tag流程(https://www.epicypher.com/resources/protocols/cutana-pag-tn5-resources/)兼容。
您可以使用现有的试剂盒组分来按照第2版的实验流程操作,而不需要任何新材料!
需要注意的主要区别是试剂盒中去掉了TAPS Wash Buffer。之前使用TAPS Wash Buffer进行标记后洗涤,现在使用等体积的Pre-Wash Buffer洗涤。EpiCypher在该试剂盒中为使用者提供了充足的Pre-Wash Buffer来完成这一步骤。
【 总 结 】
CUTANA™ CUT&Tag Kit版本的更新反映了EpiCypher严格的研发工作。EpiCypher团队将继续完善CUT&Tag和CUT&RUN的相关研究,包括针对新应用和研究领域的优化。如果您有其他疑问,第2版的实验操作流程(https://www.epicypher.com/resources/protocols/cutana-cut-and-tag-kit-manual/)也许能解决您的问题,也欢迎联系EpiCypher中国代理商上海金畔生物咨询。
【 参考文献 】
1. Janssens DH et al. Scalable single-cell profiling of chromatin modifications with sciCUT&Tag. Nat Protoc 19, 83-112 (2024). https://doi.org/10.1038/s41596-023-00905-9.
2. Bartosovic M et al. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat Biotechnol 39, 825-35 (2021). https://doi.org/10.1038/s41587-021-00869-9.
3. Wu SJ et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat Biotechnol 39, 819-24 (2021). https://www.doi.org/10.1038/s41587-021-00865-z.
4. Deng Y et al. Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 375, 681-6 (2022). https://doi.org/10.1126/science.abg7216.
5. Zhang D et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113-22 (2023). https://doi.org/10.1038/s41586-023-05795-1.
6. Henikoff S et al. Direct measurement of RNA Polymerase II hypertranscription in cancer FFPE samples. bioRxiv 2024.02.28.582647 (2024). https://doi.org/10.1101/2024.02.28.582647.
7. Henikoff S et al. Epigenomic analysis of formalin-fixed paraffin-embedded samples by CUT&Tag. Nat Commun 14, 5930 (2023). https://doi.org/10.1038/s41467-023-41666-z.
8. Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930 (2019). https://doi.org/10.1038/s41467-019-09982-5.
9. Kaya-Okur HS et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat Protoc 15, 3264-83 (2020). https://doi.org/10.1038/s41596-020-0373-x.
如需了解更多详细信息或相关产品,
请联系EpiCypher中国代理商–上海金畔生物
请联系EpiCypher中国代理商-上海金畔生物