EpiCypher® SNAP-CUTANA™ Spike-in用户指南
EpiCypher® SNAP-CUTANA™ Spike-in User Guide
This User Guide describes EpiCypher’s quantitative nucleosome spike-in technology, or SNAP (Sample Normalization and Antibody Profiling) Spike-in Controls for CUTANA™ CUT&RUN and CUT&Tag assays.
Table of Contents
1. CUTANA™ CUT&RUN and CUT&Tag Assay Overview …………………………………………….2
2. SNAP-CUTANA™ Spike-in Controls: Overview and Advantages …………………………….. 4
3. Applications of SNAP-CUTANA™ Spike-ins for CUT&RUN and CUT&Tag ………………. 6
3.1. CUT&RUN/CUT&Tag assay optimization. …………………………………………………………… 6
3.2. Monitor experimental success. …………………………………………………………………………… 7
3.3. Identify high quality histone PTM antibodies. ……………………………………………………… 10
3.4. Normalize data and quantitatively compare samples…………………………………………… 12
4. Addition of SNAP-CUTANA™ Spike-ins to CUT&RUN and CUT&Tag reactions …….. 14
4.1. Adding the K-MetStat Panel to CUT&RUN reactions …………………………………………… 14
4.2. Adding the K-MetStat Panel to CUT&Tag reactions ……………………………………………. 15
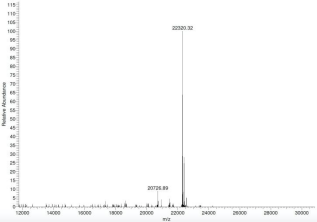
5. Analysis of SNAP-CUTANA™ Spike-in Control Sequencing Data …………………………. 16
5.1. Count the number of sequencing reads assigned to each nucleosome in the panel.... 16
5.2. Generate a heatmap of the spike-in reads …………………………………………………………. 17
5.3. Examine spike-in data ……………………………………………………………………………………… 18
6. SNAP-CUTANA™ Spike-in Panel DNA Barcodes ………………………………………………….. 19
7. References …………………………………………………………………………………………………………20
1. CUTANA™ CUT&RUN and CUT&Tag Assay Overview
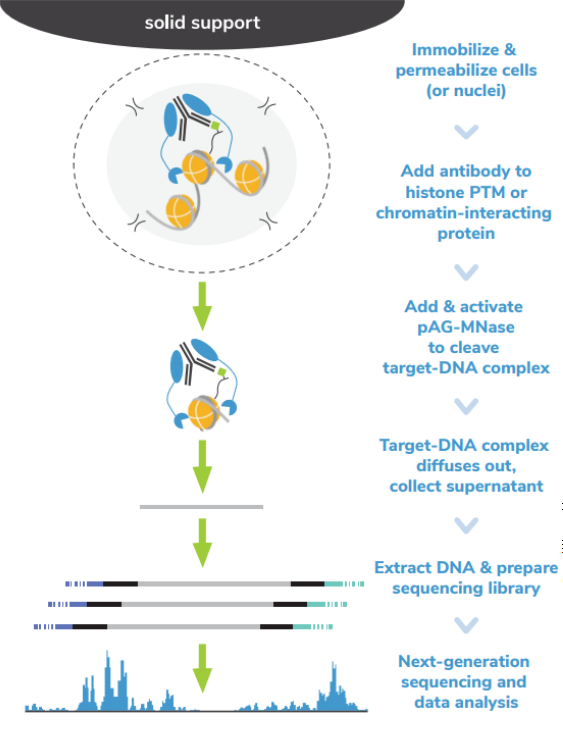
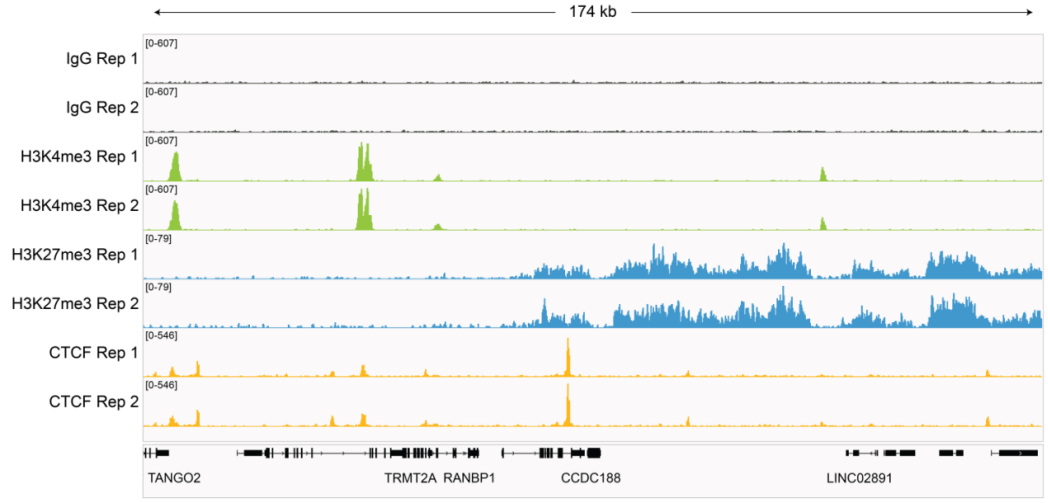
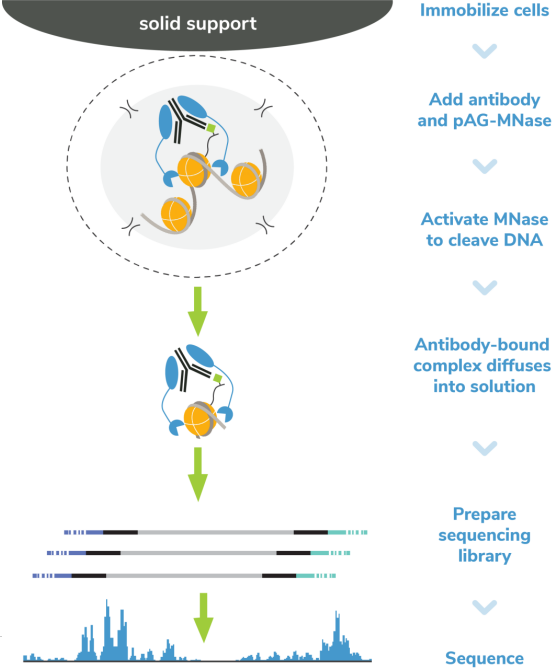
Cleavage Under Targets & Release Using Nuclease (CUT&RUN) and Cleavage Under Targets & Tagmentation (CUT&Tag) are revolutionary genomic mapping strategies developed by the group of Dr. Steven Henikoff1,2. Both assays build on the Chromatin ImmunoCleavage (ChIC) approach from Dr. Ulrich Laemmli3, in which Protein A/G is used to recruit an enzymatic domain to antibody-bound chromatin in situ3. An important feature of CUT&RUN/CUT&Tag is the immobilization of cells (or nuclei) to solid support (Figure 1), which streamlines assay workflows, improves signal-to-noise (vs. ChIP), and enables low cell inputs and sequencing requirements.
In CUT&RUN, a pAG-Micrococcal Nuclease (pAG-MNase) fusion is used to cleave antibodylabelled chromatin. Fragments are released from cells and purified (Figure 1A). In CUT&Tag, pAG is fused with prokaryotic transposase 5 (pAG-Tn5) to cleave and tagment antibody-bound chromatin with sequencing adapters (Figure 1B). Both assays are compatible with nextgeneration sequencing (NGS) to provide high quality profiles of histone post-translational modifications (PTMs) and chromatin-associated proteins. Robust CUT&RUN and CUT&Tag protocols, based on EpiCypher’s validated workflows, are available (epicypher.com/protocols).
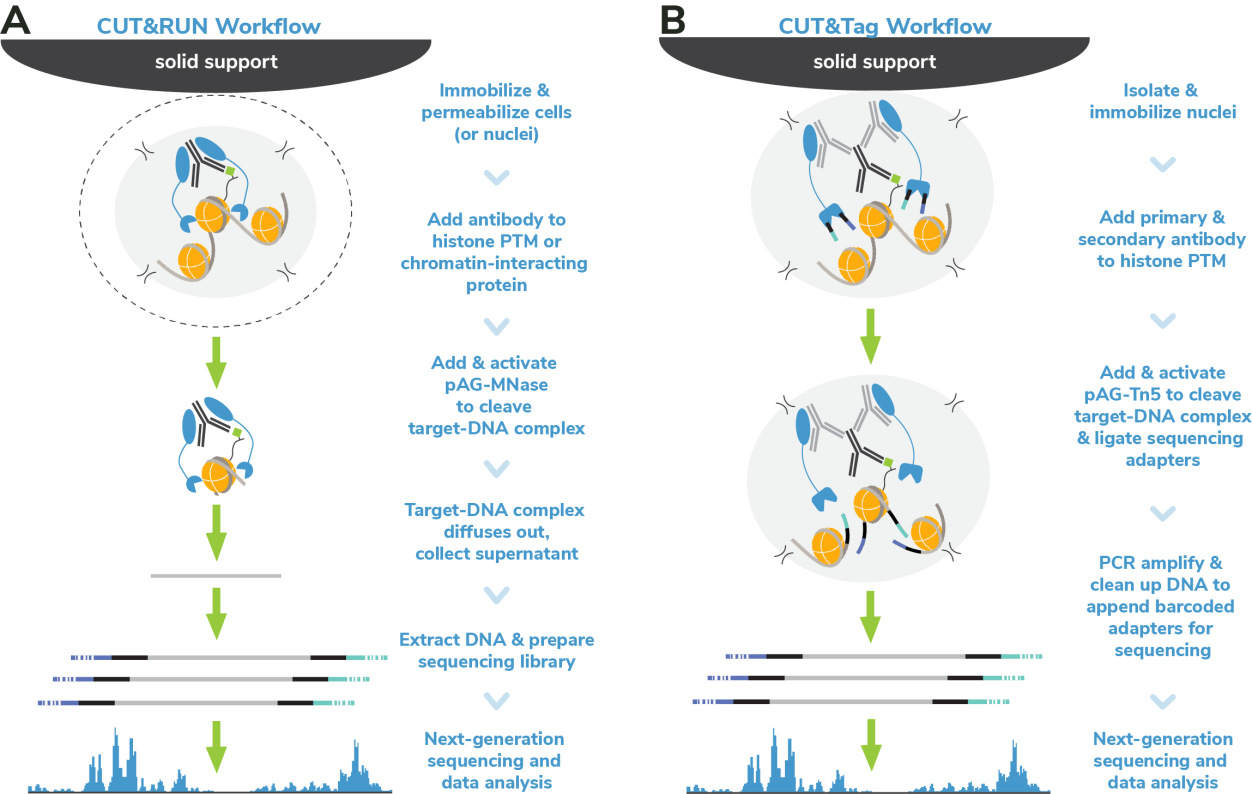
Figure 1: Overview of the CUTANA™ CUT&RUN (A) and CUT&Tag (B) protocols.
Improved controls for CUT&RUN and CUT&Tag experiments
Compared to Chromatin ImmunoPrecipitation sequencing (ChIP-seq), the leading chromatin mapping assay, CUT&RUN and CUT&Tag consistently generate higher quality data with improved signal-to-noise, while using a fraction of the cellular input and sequencing depth.
But how can users be sure these new assays work as intended? What controls are available?
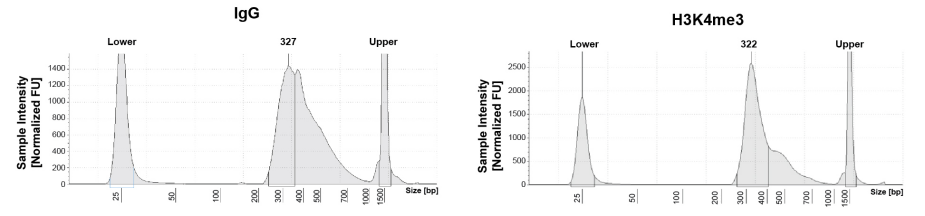
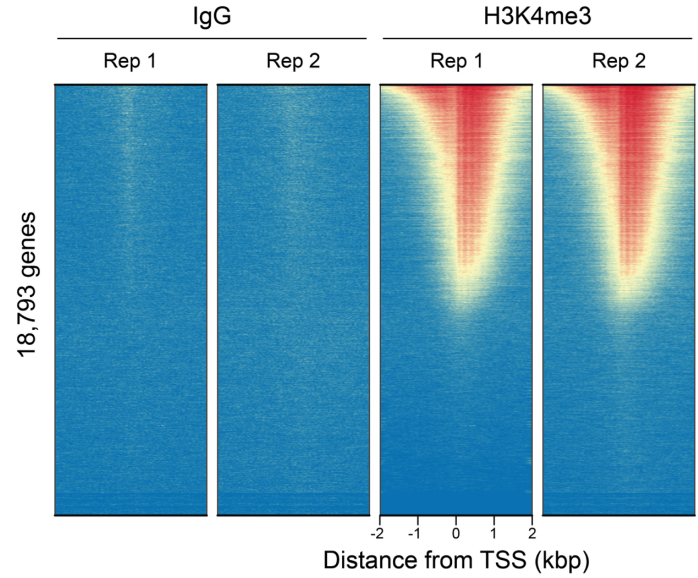
Typical CUT&RUN/CUT&Tag experiments include positive (e.g. H3K4me3) and negative (e.g. IgG) control antibodies and validated cell lines (e.g. K562 cells). For both assays we also recommend confirming cell viability and bead binding, as well as the size distribution of final NGS libraries (described in the CUTANA™ CUT&RUN and CUT&Tag protocols at epicypher.com/protocols). However, even with such robust controls, questions remain:
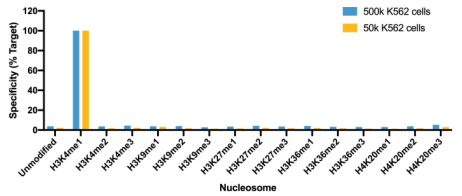
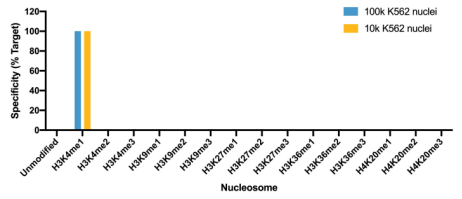
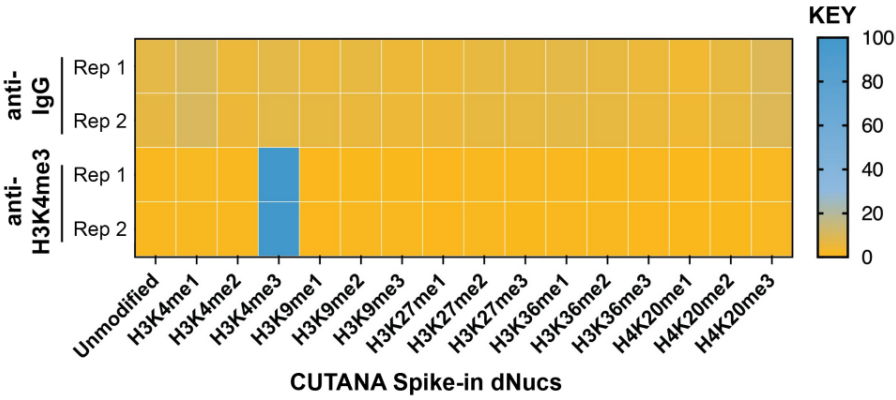
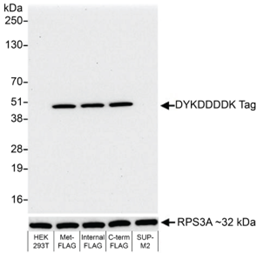
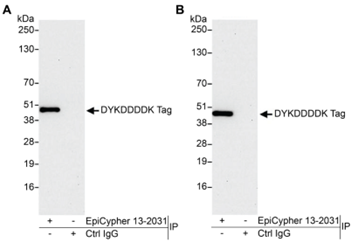
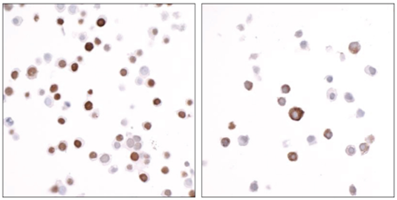
• Is my histone PTM antibody (including H3K4me3 control antibody) specific? Our previous4 and ongoing work (chromatinantibodies.com) has shown that PTM antibodies frequently cross-react with related PTM targets and can exhibit application-specific performance. Through our extensive development of CUT&RUN/CUT&Tag assays to various targets, EpiCypher has found that antibodies that work well in ChIP may not always work in CUT&RUN and/or CUT&Tag. In-application testing against a defined panel of onand off-target nucleosome substrates is the ideal strategy to identify high qualityantibodies4.
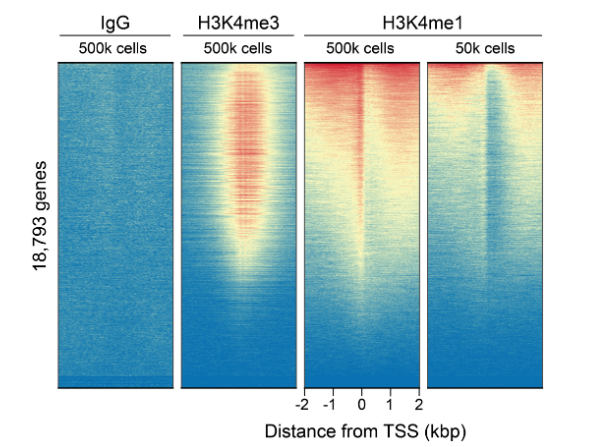
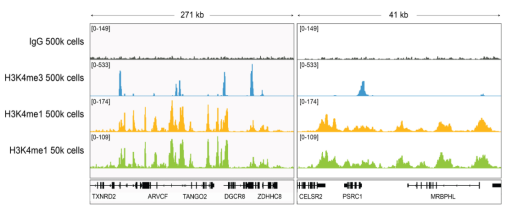
• If my reaction fails or NGS data are of poor quality, how do I troubleshoot my experiment? CUT&RUN and CUT&Tag methods can be challenging, especially for new users, and it is not always clear which step requires optimization. For instance, poor signalto- noise in NGS data could be due to failed enzymatic activity, poorly optimized buffe conditions, antibody cross-reactivity, bead clumping, or problems with cell preparation, among others. Defined spike-in controls that replicate physiological chromatin structure (i.e. nucleosomes) can help optimize workflows and guide troubleshooting strategies.
EpiCypher is a leader in the development of semi-synthetic/recombinant nucleosomes and has recently leveraged this technology for the development of SNAP (Sample Normalization and Antibody Profiling) Spike-in Controls (epicypher.com/technologies/snap-spike-in-controls). Our recently launched SNAP-CUTANA™ K-MetStat Panel can be used as a quantitative spike-in control in both CUT&RUN and CUT&Tag reactions targeting histone lysine methylation PTMs.
In this User Guide we describe SNAP-CUTANA™ Spike-ins using the K-MetStat Panel (EpiCypher #19-1002) as our primary example. We review how to incorporate these spikeins into CUT&RUN/CUT&Tag approaches, data analysis, and interpretations. For additional product information, visit epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls.
EpiCypher® SNAP-CUTANA™ Spike-in用户指南(完整版)下载链接:SNAP-CUTANA_K-MetStat_user_guide.pdf (epicypher.com),
扫描二维码下载:

如需了解更多详细信息或相关产品,请联系EpiCypher中国代理商-上海金畔生物